Predicting age irrespective of task: a modelling approach to study demographic alterations in the hive
Predicting age irrespective of task: a modelling approach to study demographic alterations in the hive
Background
Recent research proposed the existence of a common pathway to colony collapse, related to the accelerated onset of foraging (Perry et al., 2015), while another group found a common response to different pathogens involving the expression of vitellogenin, a key protein in foraging regulation (Doublet et al., 2017). To date, no methods are described to study and detect demographic alterations like those related to precocious foraging or prolonged nursing. The aim of this experiment, conducted within the project “Biomarker discovery in honey bees” funded by the Trust, is to discriminate the age of nurse and foragers through the study of the electrophoretic pattern of haemolymph proteins.
Approach
Trials were conducted on single cohort colonies (SCC), placed in a dedicated apiary near the Department of Veterinary Medical Sciences (DIMEVET) of the University of Bologna, Italy. SCCs are colonies made with a variable number of same aged workers and a fertile queen. Two trials with the same setup were conducted in June-July and September-October. In each trial, nurses and foragers were sampled. Combining the task in the hive with the known age, four categories of bees were sampled: correct aged foragers (subsequently referred as “correct_for”, n=35), precocious foragers (subsequently referred as “precoc_for”, n=28), correct aged nurses (subsequently referred as “correct_nur”, n=35), over aged nurses (subsequently referred as “over_nur”, n=36).

Figure 1 - Collection of nurse bees on the combs
Two microliters of haemolymph were drawn for each bee with a graduated glass microcapillaryand the protein separated via SDS-PAGE. The gels were stained with Coomassie G250, digitalised and manipulated with the software Fiji, based on ImageJ 1.52i (Schindelin et al., 2012). At the end of the analysis, a database was created with columns representing the samples, rows representing distances from the loading well (about 100 distances, corresponding to a resolution of a value every 0.65 mm) and at the intersection of the two the intensities of the pixels. The statistical analysis was carried out with the statistical software R 3.5.2, and the RStudio IDE.
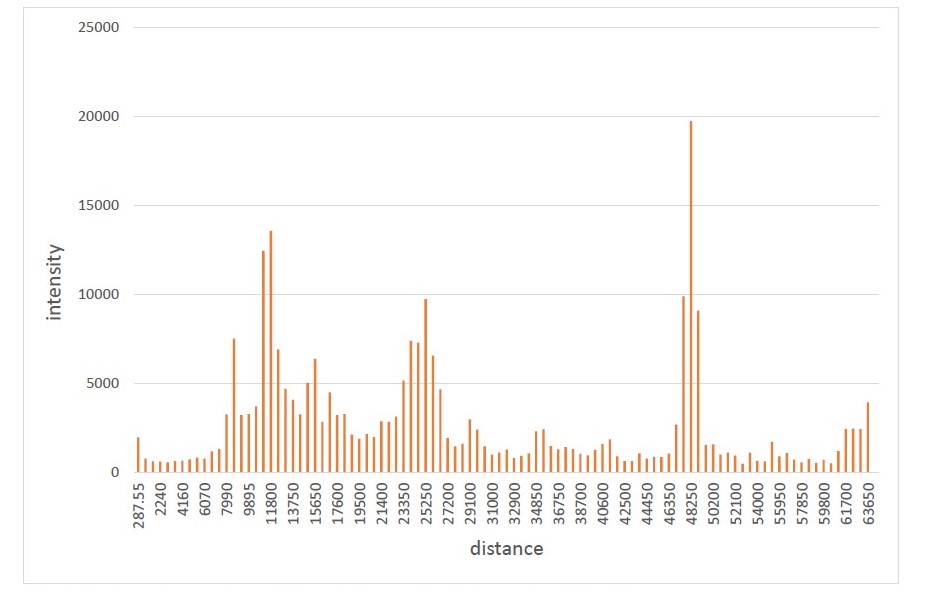
Figure 2 - Electropherogram of one of the samples (109)
A supervised learning approach was followed to create three different models: correct_for / correct_nur, correct_nur / over_nur, precoc_for / correct_for. Each dataset was randomly split in two: train (80% of the cases) and test (20% of the cases). On the test dataset variables were selected trough RKNN-FS(Li et al., 2011), while the train dataset was used to fit two different models: pls (Partial Least Squares regression) and kNN (K-nearest neighbours). Parameters and performances were assessed on train dataset trough a k-fold cross-validation procedure, consisting of 10 folds and 5 repeats. Performances of each model were assessed through ROC, Sensibility, Specificity and AUC (Area Under the Curve).
Findings
The models fitted with the values for correct_nur and correct_for gave the best results among the three. Different ages and different tasks lead to a good predictive accuracy as summarized by the AUC value. The PLS model performed significantly better.
|
|
PLS (ncomp=3) |
kNN (k=9) |
|
ROC |
0.90 |
0.91 |
|
Sens. |
0.81 |
0.80 |
|
Spec. |
0.75 |
0.70 |
|
AUC |
0.87 |
0.62 |
Table 1 - Summary of the models fitted to the dataset containing correct aged nurses and foragers
The models fitted with the dataset containing data from correct_nur and over_nur performed worse compared to the previous ones. The PLS model performed significantly better than the kNN in this case as well.
|
nur |
PLS (ncomp=5) |
kNN (k=9) |
|
ROC |
0.82 |
0.78 |
|
Sens. |
0.66 |
0.62 |
|
Spec. |
0.77 |
0.84 |
|
AUC |
0.78 |
0.60 |
Table 2 - Summary of the models fitted to the dataset containing correct aged and over aged nurses
Lastly, the prediction between precoc_for and correct_for, is the most cumbersome. In this case the kNN model performed no better than random guessing while PLS model performed slightly better.
|
for |
PLS (ncomp=6) |
kNN (k=5) |
|
ROC |
0.70 |
0.63 |
|
Sens. |
0.75 |
0.84 |
|
Spec. |
0.48 |
0.36 |
|
AUC |
0.64 |
0.49 |
.
Table 3 - Summary of the models fitted to the dataset containing correct aged and precocious foragers
Conclusions
The first model studied gave, as expected, very good results. However, its function in the study was only to validate the approach, testing it on individuals with very different physiological traits and thus presumably easy to discriminate. The PLS model fitted with the nurses dataset showed a good predictive accuracy. Climate change and intensive agriculture are shaping colony population dynamics. During periods of brood shrinkage, the lack of newly emerged bees probably leads them to an extended time of brood caring to compensate. Moreover, the same situation is likely found after natural and artificial brood interruption. As highlighted in the work of Wegener et al. (2009), overaged nurses are not functionally equivalent to young ones; the degeneration of mandibular glands leads to phenotypic differences in the reared workers which exhibit higher ovary development. The application of such model is relevant to study these phenomena and potentially counteract them. Unfortunately, none of the models fitted for foragers exhibited enough predictive accuracy. The sample size, though not scarce, it’s on the low side for this type of approach; more samples means more variables retainable and possibly a better and usable model. So, more work needs to be done to refine the models and test them on full sized colonies in the field.
Riccardo Cabbri
Bologna University
The grant from the Eva Crane Trust supported my PhD project (Biomarker discovery in honey bees) of which this experiment is part. The funding allowed me to work on other trials that got published this year (Cabbri et al., 2018[1] and Sgolastra et al., 2018) and to present two posters at the Eurbee meeting that took place in Ghent (Cabbri et al., 2018 [2], Cabbri et al., 2018 [3]).
References
Doublet, V., Poeschl, Y., Gogol-Döring, A., Alaux, C., Annoscia, D., Aurori, C., ... & Flenniken, M. L. (2017). Unity in defence: honeybee workers exhibit conserved molecular responses to diverse pathogens. BMC genomics, 18(1), 207.
Li, S., Harner, E. J., & Adjeroh, D. A. (2011). Random KNN feature selection-a fast and stable alternative to Random Forests. BMC bioinformatics, 12(1), 450.
Perry, C. J., Søvik, E., Myerscough, M. R., & Barron, A. B. (2015). Rapid behavioral maturation accelerates failure of stressed honey bee colonies. Proceedings of the National Academy of Sciences, 112(11), 3427-3432.
Schindelin, J.; Arganda-Carreras, I. & Frise, E. et al. (2012), "Fiji: an open-source platform for biological-image analysis", Nature methods 9(7): 676-682, PMID 22743772, doi:10.1038/nmeth.2019
Wegener, J., Lorenz, M. W., & Bienefeld, K. (2009). Physiological consequences of prolonged nursing in the honey bee. Insectes Sociaux, 56(1), 85-93.
