Background
Broadcast spray application of antibiotics in crop fields is increasing exponentially in the United States in response to expanding bacterial agricultural pathogens. Yet we have almost no understanding of how these mass field antibiotic applications will affect beneficial organisms, including pollinators. This is concerning because laboratory evidence indicates that bee gut symbionts can influence bee learning (Zhang et al., 2020; Li et al., 2021), and that antibiotics used to treat pathogens within hives can impact bee foraging (Zhang et al., 2020; Ortiz-Alvarado et al., 2021). Declines in learning and foraging could affect colony growth and pollination.
Research
In this project, we contribute to this burgeoning field of research by testing the effect of an upper-limit dietary exposure to streptomycin, an antibiotic widely used in orchards in the United States, on Bombus impatiens, a wild bumble bee that is also commercially available for crop pollination. We found that feeding a sucrose solution with 200 ppm of streptomycin for 48 hrs negatively impacted bumble bee learning and foraging (Figure 1 A & B). Additionally, contrary to expectations, the microbial diversity in the bee gut increased (Figure 1 C) due to a decline in the abundance of core bacterial taxa and the proliferation of opportunistic bacteria (Figure 2).
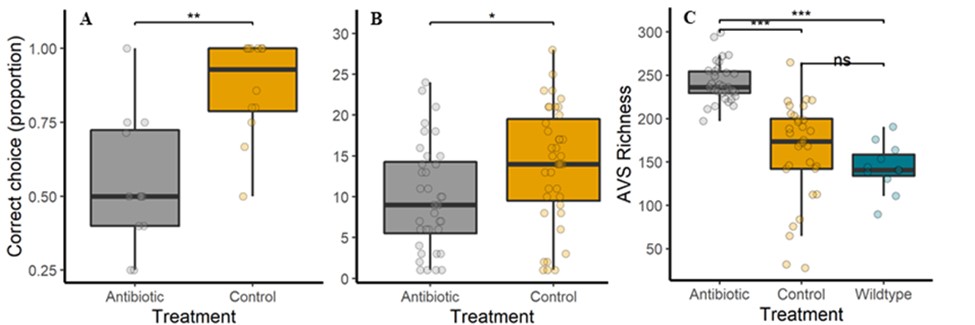
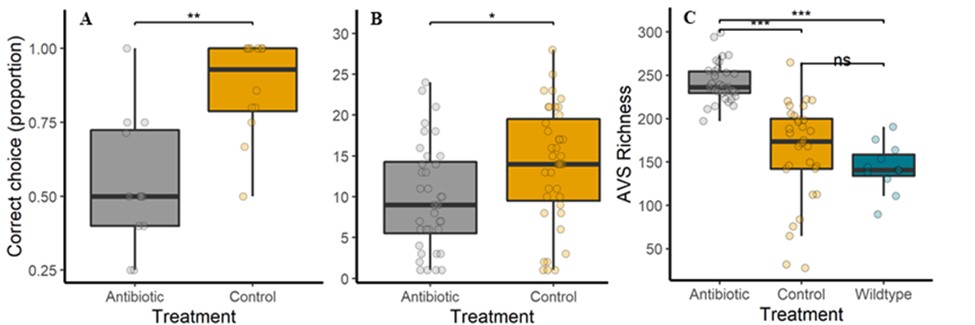
Figure 1. After exposure to 200 ppm streptomycin vs. a control sucrose diet, foragers are less likely to A) make the correct choice in an associative learning test (n=102 bees, five colonies, Wald X2=18.82, P<0.001) and to B) feed on a sucrose reward (n=95 bees, five colonies, Wald X2=5.63, P<0.05). Furthermore, C) gut bacterial richness (Amplicon Sequence Variant) increases as the core symbiont abundance decreases, allowing rare taxa to proliferate: antibiotic (n=31 bees), sucrose control (n=30 bees), wildtype/untreated (n=11 bees). Asterisks = P<0.001 in a Tukey-Bonferroni test. These data are currently in review for publication.
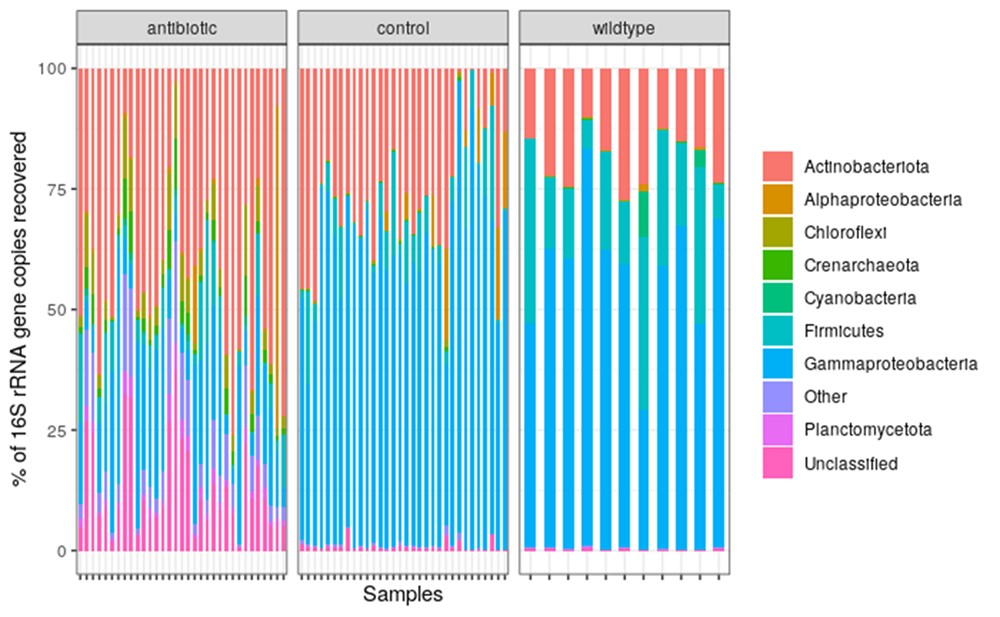
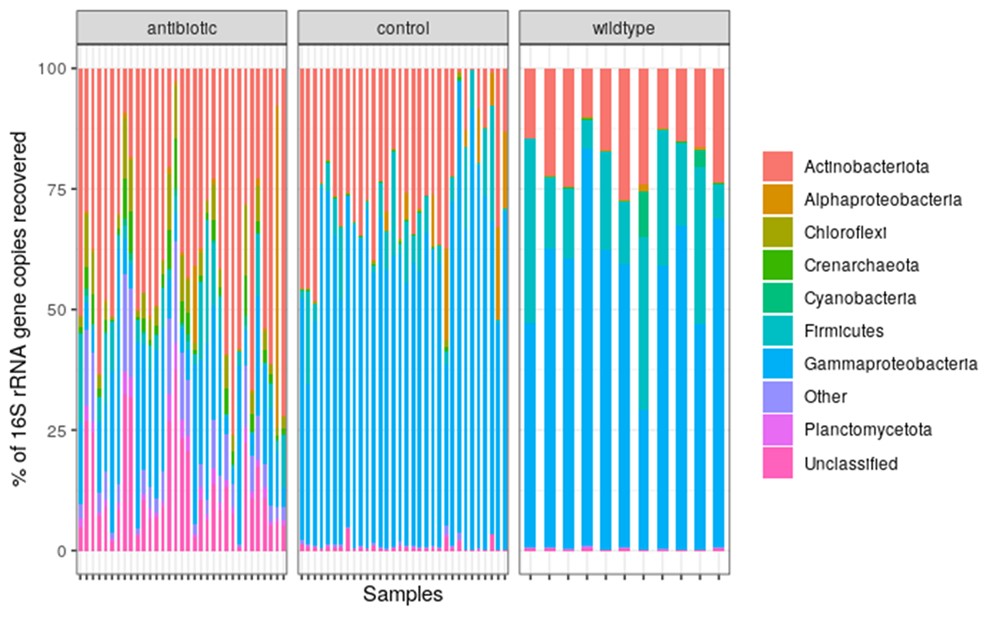
Figure 2. Relative abundance of major Phyla present across antibiotic-treated (n=31), control (n=30), and wildtype/untreated (n=11) bumble bees (Bombus impatiens).
In addition, we cultured bacteria from honey bee guts collected at two apple orchards (Georgia, USA) that received antibiotic applications (streptomycin and oxytetracycline) and a control strawberry site (without antibiotics). We found statistically significant differences in the cultured microbial communities from bee guts at both locations (Figure 3). However, we did not find statistically significant differences in the proportion of those bacteria resistant to streptomycin in-vitro (Figure 4). Further, we found that strA-strB, an antibiotic-resistant gene previously reported by others (Tian et al., 2012; Ludvigsen et al., 2018), was also present in bees across sites, indicating that resistance to streptomycin might be more common than expected in this region (Figure 5). We are awaiting sequencing data to investigate longitudinal changes in sentinel honey bee and bumble bee microbiome composition before, during, and after antibiotic exposure at apple orchards. In addition we are optimizing the quantification of antibiotic residues on pollen, flowers and bees via ELISA tests.
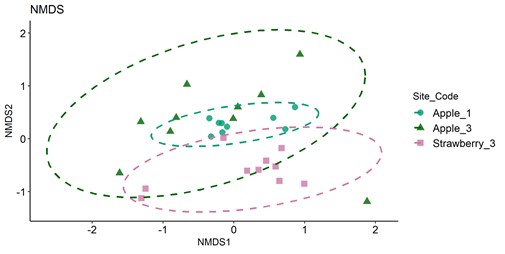 Figure 3. Non-Metric Multidimensional Scaling (NMDS) of in-vitro bacterial communities from honey bees. We sampled honey bee foraging on apple flowers (that had received antibiotic treatments) in two orchards, and strawberry flowers (no antibiotic applications) at one site. We dissected and plated the bee’s guts in four types of agar media. We later performed direct PCR of bacterial colonies to amplify the 16s rDNA gene for posterior Sanger sequencing and taxonomical assignment. We used a Bray-Curtis similarity matrix to estimate a PERMANOVA (‘adonis’ function from the ‘vegan’ package). There were statistically significant differences among bees sampled at apple orchards vs. the strawberry site (P=0.0001). Each point, square, and triangle represents the bacterial community of a single bee.
Figure 3. Non-Metric Multidimensional Scaling (NMDS) of in-vitro bacterial communities from honey bees. We sampled honey bee foraging on apple flowers (that had received antibiotic treatments) in two orchards, and strawberry flowers (no antibiotic applications) at one site. We dissected and plated the bee’s guts in four types of agar media. We later performed direct PCR of bacterial colonies to amplify the 16s rDNA gene for posterior Sanger sequencing and taxonomical assignment. We used a Bray-Curtis similarity matrix to estimate a PERMANOVA (‘adonis’ function from the ‘vegan’ package). There were statistically significant differences among bees sampled at apple orchards vs. the strawberry site (P=0.0001). Each point, square, and triangle represents the bacterial community of a single bee.
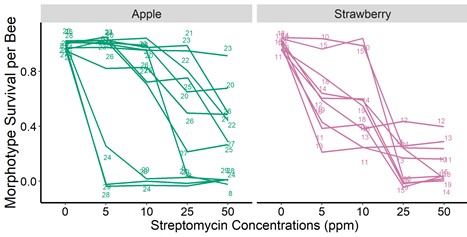
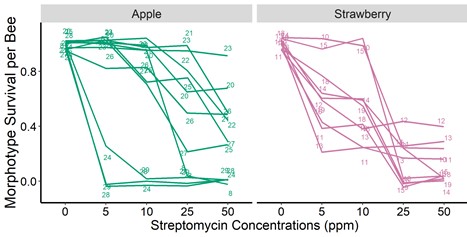 Figure 4. The proportion of bacteria recovered from the bee guts (indicated with numbered lines) that survived increasing streptomycin concentrations in-vitro. We dissected the bee guts following standard protocols, homogenized the entire gut in a saline solution, and plated the guts in four different agar media. We incubated the agar plates at 37 Celsius for 48 hrs in a regular atmosphere and CO2 enriched incubator. We did not find statistically significant differences among treatments.
Figure 4. The proportion of bacteria recovered from the bee guts (indicated with numbered lines) that survived increasing streptomycin concentrations in-vitro. We dissected the bee guts following standard protocols, homogenized the entire gut in a saline solution, and plated the guts in four different agar media. We incubated the agar plates at 37 Celsius for 48 hrs in a regular atmosphere and CO2 enriched incubator. We did not find statistically significant differences among treatments.
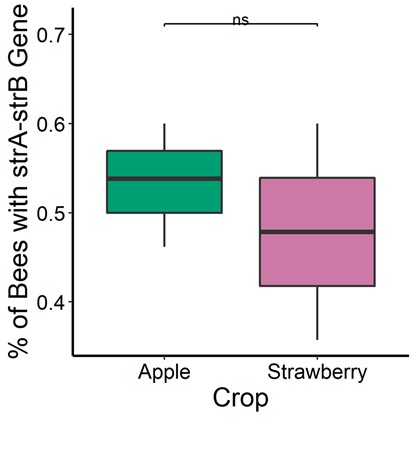
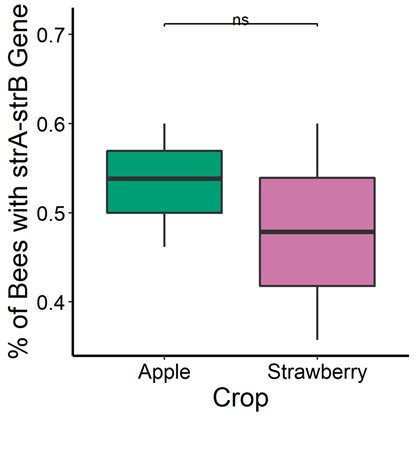
Figure 5. Prevalence of streptomycin (strA-strB resistance). Honey bee foragers were sampled at three apple orchards (n= 66 bees ) and two strawberry sites (n=29 bees) sites. Microbes from bee guts were equally likely to carry the strA-strB gene regardless of the source crop (binomial regression for individual bees, P = 0.4027).
Summary
We found negative impacts of an upper-limit streptomycin dose on bumble bee learning and foraging, and widespread streptomycin resistance in honey bees in our region. These findings further our understanding of the impact of agricultural antibiotics on bees’ microbiome and foraging behavior. Still, much work remains to characterize field-realistic exposure to antibiotic residues in crop nectar and pollen.
Laura Avila, Ph.D. - Emory University, Georgia, USA
Berry J. Brosi, Ph.D. - University of Washington, Washington, USA
Ref.: ECTA_20191220
Completed: 2022
Acknowledgments: We are very grateful for the support of the Eva Crane Trust. We also thank personnel at the Georgia Mountain Research Station of the University of Georgia and extension agents associated with the University of Georgia for facilitating our field work. We thank numerous undergraduate research assistants and Dr. Nicole Gerardo for guidance and laboratory space.
This work has been presented at the following conferences:
Avila, L., Griffin, B., Gray, A., McGrath, Balu, T., A., B. J. Brosi, and N.M. Gerardo. (2021, Oct 31-Nov 3). Assessing the impact of broadcast-sprayed antibiotics on bee gut microbiome composition and presence of antibiotic-resistant genes. Entomological Society of America Annual Meeting, online conference.
Avila, L., Gerardo, N.M., and B. J. Brosi. (2021, Oct 18-20). The impact of crop prophylactic bactericides on bee microbiome, foraging behavior, and pollination. North American Pollinator Protection Campaign Annual Meeting.
Avila, L., Perrin, S., McGrath, A., Griffin, B., Hofmann, D., Boyd, S., Covington, R., Blaauw, B.R., Gerardo, N., and Brosi, B.J. (2020, Nov 11-25). The impact of crop prophylactic bactericides on bee microbiome, foraging behavior, and pollination. Entomological Society of America Annual Meeting, online conference. 1 page.
Avila, L., Dunne L., and B. J. Brosi. 2020. Managed bumble bees alter their foraging behavior when fed agricultural antibiotics. Ecological Society of America Annual Meeting, Online. Poster.
References Cited
Li L, Solvi C, Zhang F, Qi Z, Chittka L, Zhao W. 2021 Gut microbiome drives individual memory variation in bumblebees. Nat. Commun. 2021 121 12, 1–10. (doi:10.1038/s41467-021-26833-4)
Ludvigsen, J., Amdam, G. V., Rudi, K. & L’Abée-Lund, T. M. 2018 Detection and Characterization of Streptomycin Resistance (strA-strB) in a Honeybee Gut Symbiont (Snodgrassella alvi) and the Associated Risk of Antibiotic Resistance Transfer. Microb. Ecol. 76, 588–591
Muth F, Cooper TR, Bonilla RF, Leonard AS. 2018 A novel protocol for studying bee cognition in the wild. Methods Ecol. Evol. 9, 78–87. (doi:10.1111/2041-210X.12852)
Ortiz-Alvarado Y, Clark DR, Vega-Melendez CJ, Flores-Cruz Z, Domingez-Bello MG, Giray T. 2020 Antibiotics in hives and their effects on honey bee physiology and behavioral development. Biol. Open 9. (doi:10.1242/BIO.053884)
Tian, B., Fadhil, N. H., Powell, J. E., Kwong, W. K. & Moran, N. A. 2012 Long-Term Exposure to Antibiotics Has Caused Accumulation of Resistance Determinants in the Gut Microbiota of Honeybees. MBio 3, e00377-12
Zhang Z, Mu X, Cao Q, Shi Y, Hu X, bioRxiv HZ-, 2020 undefined. 2020 Honeybee gut microbiota modulates host behaviors and neurological processes. biorxiv.org (doi:10.1101/2020.12.19.423587)

